The Beginner’s Guide to R Packages
Now that we’ve
installed R and R Studio and have a basic understanding of how they work
together, we can get at what makes R so special: packages.
What is an R package?
So far, anything we’ve
played around with in R uses the “base” R system. Base R, or everything
included in R when you download it, has rather basic functionality for
statistics and plotting but it can sometimes be limiting. To expand upon R’s
basic functionality, people have developed packages. A package
is a collection of functions, data, and code conveniently provided in a nice,
complete format for you. At the time of writing, there are just over 19,266 packages available to download at CRAN- each with their own specialized functions and
code, all for some different purpose. For a really in depth look at R Packages
(what they are, how to develop them), check out Hadley Wickham’s book from
O’Reilly, “R
Packages.” Link will be provided
in our blog post, so definitely heck that and please follow our blogpost too.
A package is not to be
confused with a library (these two terms are often conflated
in colloquial speech about R). A library is the place where the package is
located on your computer. To think of an analogy, a library is, well, a
library… and a package is a book within the library. The library is where the
books/packages are located.
Packages are what make
R so unique. Not only does base R have some great functionality but these
packages greatly expand its functionality. And perhaps most special of all,
each package is developed and published by the R community at large and
deposited in repositories.
What are repositories?
A repository is a
central location where many developed packages are located and available for
download.
There are three big
repositories:
1. CRAN
(Comprehensive R Archive Network): R’s main repository (>19,000 packages available!)
2. BioConductor: A repository mainly for
bioinformatic-focused packages
3. GitHub: A very popular, open source repository
(not R specific!)
Take a second to explore the links above and check out the various packages that are out there!
How do you know what package is right for you?
So you know where to
find packages… but there are so many of them, how can you find a package that
will do what you are trying to do in R? There are a few different avenues for
exploring packages.
First, CRAN groups all
of its packages by their functionality/topic into 35 “themes.” It calls this
its “Task view.” This at least allows you to narrow the packages you can
look through to a topic relevant to your interests.
Second, there is a
great website, RDocumentation, which
is a search engine for packages and functions from CRAN, BioConductor, and
GitHub (i.e.: the big three repositories). If you have a task in mind, this is a
great way to search for specific packages to help you accomplish that task! It
also has a “task” view like CRAN, that allows you to browse themes.
More often, if you
have a specific task in mind, Googling that task followed by “R package” is a
great place to start! From there, looking at tutorials, vignettes, and forums
for people already doing what you want to do is a great way to find relevant
packages.
How do you install packages?
Great! You’ve found a
package you want… How do you install it?
Installing from CRAN
If you are installing from the CRAN repository, use the install.packages() function, with the name of the package you want to install in quotes between the parentheses (note: you can use either single or double quotes). For example, if you want to install the package “ggplot2”, you would use: install.packages("ggplot2")Try doing so in your R
console! This command downloads the “ggplot2” package from CRAN and installs it
onto your computer.
If you want to install
multiple packages at once, you can do so by using a character vector,
like: install.packages(c("ggplot2",
"devtools", "lme4"))
If you want to use
RStudio’s graphical interface to install packages, go to the Tools menu, and
the first option should be “Install packages…” If installing from CRAN, select
it as the repository and type the desired packages in the appropriate box.
Installing from Bioconductor
The BioConductor repository uses their own method to install packages. First, to get the basic functions required to install through BioConductor, use: source("https://bioconductor.org/biocLite.R")This makes the main
install function of BioConductor, biocLite(), available to you. Following this, you call
the package you want to install in quotes, between the parentheses of the biocLite command, like
so: biocLite("GenomicFeatures")
Installing from GitHub
This is a more specific case that you probably won’t run into too often. In the event you want to do this, you first must find the package you want on GitHub and take note of both the package name AND the author of the package. Check out this guide for installing from GitHub, but the general workflow is:- install.packages("devtools") - only run this if you don’t already have
devtools installed. If you’ve been following along with this lesson, you
may have installed it when we were practicing installations using the R
console
- library(devtools) - more on what this command is doing immediately
below this
- install_github("author/package") replacing “author” and “package” with their
GitHub username and the name of the package.
Loading packages
Installing a package
does not make its functions immediately available to you. First you must load the
package into R; to do so, use the library() function. Think of this like any other
software you install on your computer. Just because you’ve installed a
program, doesn’t mean it’s automatically running - you have to open the
program. Same with R. You’ve installed it, but now you have to “open” it. For
example, to “open” the “ggplot2” package, you would run: library(ggplot2)
NOTE: Do not put the package
name in quotes! Unlike when you are installing the packages, the library() command does not
accept package names in quotes!
Step one of getting a package is installing it, but to use it, you must load it using library(); similar to installing R and then loading it by opening the .exe file
There is an order to
loading packages - some packages require other packages to be loaded first (dependencies).
That package’s manual/help pages will help you out in finding that order, if
they are picky.
If you want to load a
package using the R Studio interface, in the lower right quadrant there is a tab
called “Packages” that lists out all of the packages and a brief description,
as well as the version number, of all of the packages you have installed. To
load a package just click on the checkbox beside the package name
Updating, removing, unloading packages
Once you’ve got a
package, there are a few things you might need to know how to do:
Checking what packages you have installed
If you aren’t sure if you’ve already installed a package, or want to check what packages are installed, you can use either of: installed.packages() or library() with nothing between the parentheses to check!In R Studio, that
package tab introduced earlier is another way to look at all of the packages
you have installed.
Updating packages
You can check what
packages need an update with a call to the function old.packages() This
will identify all packages that have been updated since you installed them/last
updated them.
To update all
packages, use update.packages(). If you only want to update a specific
package, just use once again install.packages("packagename")
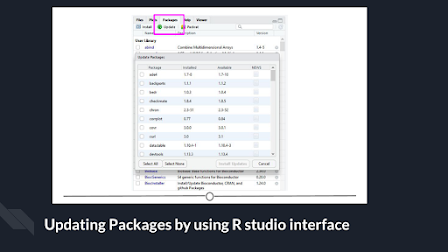
Within the R Studio
interface, still in that Packages tab, you can click “Update,” which will list
all of the packages that are not up to date. It gives you the option to update
all of your packages, or allows you to select specific packages.
You will want to
periodically check in on your packages and check if you’ve fallen out of date -
be careful though! Sometimes an update can change the functionality of certain
functions, so if you re-run some old code, the command may be changed or
perhaps even outright gone and you will need to update your code too!
Unloading packages
Sometimes you want to
unload a package in the middle of a script - the package you have loaded may
not play nicely with another package you want to use.
To unload a given
package you can use the detach() function. For example, detach("package:ggplot2", unload=TRUE) would unload the ggplot2 package (that
we loaded earlier). Within the R Studio interface, in the Packages tab, you can
simply unload a package by unchecking the box beside the package name.
Uninstalling packages
If you no longer want
to have a package installed, you can simply uninstall it using the
function remove.packages(). For example, remove.packages("ggplot2")
(Try that, but then
actually re-install the ggplot2 package - it’s a super useful plotting
package!)
Within R Studio, in the
Packages tab, clicking on the “X” at the end of a package’s row will uninstall
that package.
How do you know what version of R you have?
Sometimes, when you
are looking at a package that you might want to install, you will see that it
requires a certain version of R to run. To know if you can use that package,
you need to know what version of R you are running!
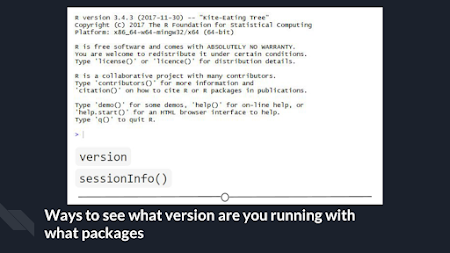
One way to know your R
version is to check when you first open R/R Studio - the first thing it outputs
in the console tells you what version of R is currently running. If you didn’t
pay attention at the beginning, you can type version into the console
and it will output information on the R version you are running. Another
helpful command is sessionInfo() - it will tell you what version of R you
are running along with a listing of all of the packages you have loaded. The
output of this command is a great detail to include when posting a question to
forums - it tells potential helpers a lot of information about your OS, R, and
the packages (plus their version numbers!) that you are using.
Using the commands in a function
In all of this
information about packages, we haven’t actually discussed how to use a
package’s functions!
First, you need to
know what functions are included within a package. To do this, you can look at
the man/help pages included in all (well-made) packages. In the console, you
can use the help() function to access a package’s help
files. Try help(package =
"ggplot2") and you will see
all of the many functions that ggplot2 provides. Within the
R Studio interface, you can access the help files through the Packages tab
(again) - clicking on any package name should open up the associated help files
in the “Help” tab, found in that same quadrant, beside the Packages tab.
Clicking on any one of these help pages will take you to that functions help page,
that tells you what that function is for and how to use it.
Once you know what
function within a package you want to use, you simply call it in the console
like any other function we’ve been using throughout this lesson. Once a package
has been loaded, it is as if it were a part of the base R functionality.
If you still have
questions about what functions within a package are right for you or how to use
them, many packages include “vignettes.” These are extended
help files, that include an overview of the package and its functions, but
often they go the extra mile and include detailed examples of how to use the
functions in plain words that you can follow along with to see how to use the
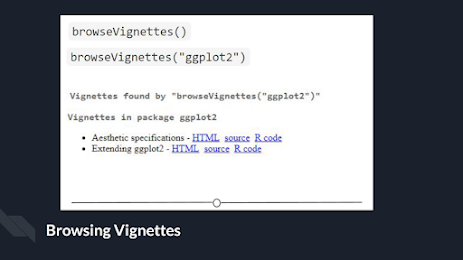
package. To see the vignettes included in a package, you can use the browseVignettes() function. For example, let’s look at the vignettes
included in ggplot2:browseVignettes("ggplot2") . You should see that there are two
included vignettes: “Extending ggplot2” and “Aesthetic specifications.”
Exploring the Aesthetic specifications vignette is a great example of how
vignettes can be helpful, clear instructions on how to use the included
functions.
Summary
In this lesson, we’ve
explored R packages in depth. We examined what a packages is (and how it
differs from a library), what repositories are, and how to find a package
relevant to your interests. We investigated all aspects of how packages work:
how to install them (from the various repositories), how to load them, how to
check which packages are installed, and how to update, uninstall, and unload
packages. We took a small detour and looked at how to check what version of R
you have, which is often an important detail to know when installing packages.
And finally, we spent some time learning how to explore help files and
vignettes, which often give you a good idea of how to use a package and all of
its functions.
If you still want to learn more about R packages, here are two great resources! R Packages: A Beginner’s Guide from Adolfo Álvarez on DataCamp and a lesson from the University of Washington, on an Introduction to R Packages from Ken Rice and Timothy Thornton. I’ll provide those links on my blogpost for this lecture.











Comments
Post a Comment
Type your comment here.
However, Comments for this blog are held for moderation before they are published to blog.
Thanks!